Over 440 SCI articles published! BGI Genomics cancer genetics and cell research fuels innovation
2022-11-17
BGI Genomics has currently published over 440 SCI (Science Citation Index) papers in the field of cancer research, with a cumulative impact factor of over 4,000 points. A consistent number of important BGI Genomics papers have been published in Nature Medicine, Cell, Nature, Science, Cell Research, Nature Genetics and other top scientific journals, including more than 40 papers with scores above 30 and nearly 70 papers with scores above 10.
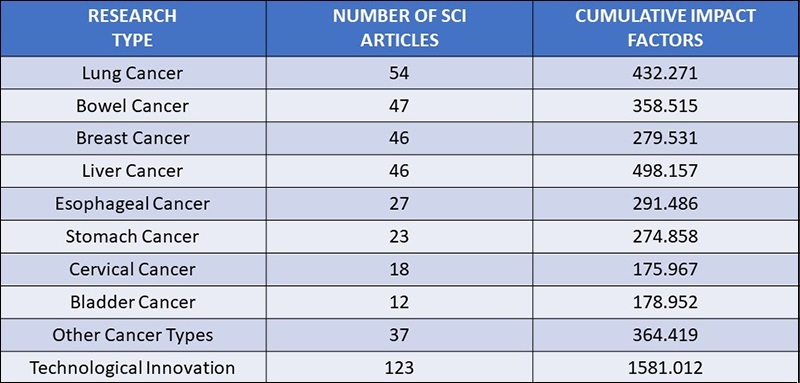
BGI Genomics research scope covers 20 types of cancers, including lung cancer, esophageal cancer, colorectal cancer, gastric cancer, pancreatic cancer, liver cancer, breast cancer, cervical cancer, endometrial cancer, nasopharyngeal cancer, thyroid cancer, prostate cancer, lymphoma, melanoma, glioma, and brain tumors.
As we have a lot of material to cover, we will introduce these BGI Genomics published scientific results in a few articles. Today, we start with a quick look at six high-impact pieces of research.

Panoramic immune profiling of early recurrent hepatocellular carcinoma using single-cell technology
Publication date: January 21st, 2021
Publication: Cell IF 66.850
Research results: Hepatocellular carcinoma (HCC) has a high recurrence rate and a low, 5-year survival rate. The team used single-cell sequencing to analyze the transcriptome information of approximately 17,000 cells from 18 primary or early recurrent HCC cases. Grouped in two separate cohorts, early recurrent tumors showed reduced levels of regulatory T cells, increased levels of dendritic cells (DCs) and infiltrating CD8+ T cells, compared to primary tumors. Notably, CD8+ T cells differed from the classical depleted state observed in primary HCC and were instead characterized by high expression of KLRB1 (CD161) in an innate-like, low-killing, and low clonal expansion state. The increase of these cells was associated with a poorer HCC prognosis. The team also revealed a potential immune evasion mechanism in recurrent tumor cells that inhibits DC antigen presentation and recruits innate CD8+ T cells, providing more insight and a more reliable basis for a comprehensive understanding of the immune evasion mechanisms associated with tumor recurrence in the HCC ecosystem.
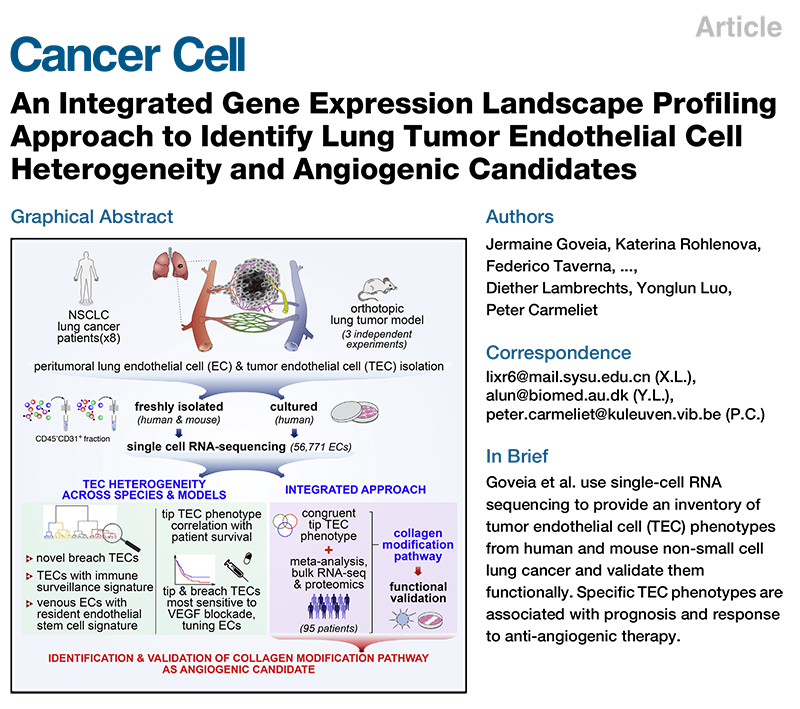
Comprehensive gene expression profiling identifies potential pathways of endothelial cell heterogeneity and angiogenesis in lung tumors
Published: January 13, 2020
Publication: Cancer Cell IF 38.585
Research results: At the single-cell level, there is a paucity of studies on the heterogeneity of lung tumor endothelial cell (TEC) phenotypes among patients, species (human/mouse), and models (in vivo/in vitro). The team sequenced single-cell RNA (scRNA) from 56,771 human/mouse lung tumor endothelial cells and cultured human lung TECs, identified 17 known and 16 previously unknown phenotypes, including TECs regulating immune surveillance function. We resolved the canonical tip TECs into a known migratory tip and the other category with a basement membrane plasticity breakthrough phenotype (tip/breach TECs). Of these 2, the tip TEC expression profile correlates with patient survival, while the tip/breach TEC phenotype is most sensitive to vascular endothelial growth factor inhibitors. Among the various TEC phenotypes, only tip TEC has cross-species, cross-model consistency and co-expresses conserved marker genes. After integrating and analyzing single-cell transcriptome sequencing data, independent bulk multi-omics data and meta-analysis data from different human tumors and validating them with functional analysis, the researchers identified collagen modification as a novel angiogenic candidate pathway.
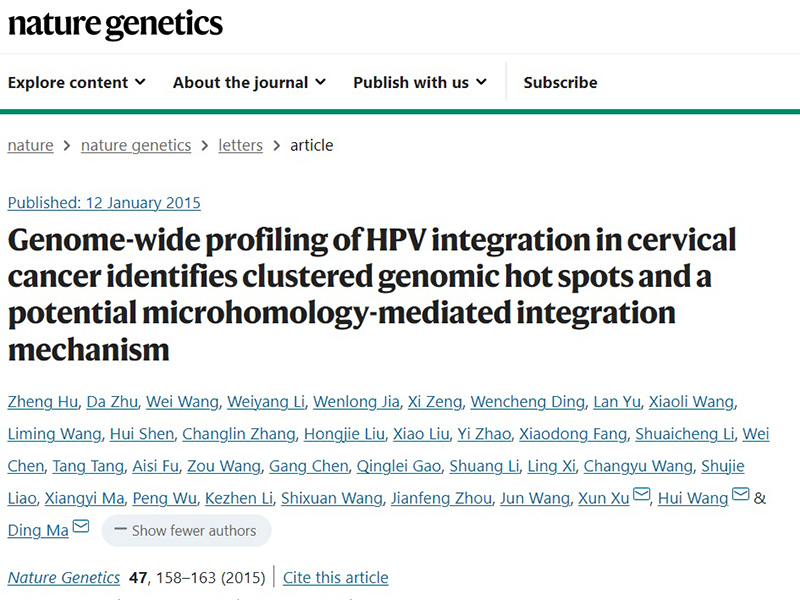
Genome-wide profiling of HPV integration in cervical cancer identifies oncogenic hotspots and potential mechanisms of oncogenesis
Published: January 12, 2015
Impact factor: Nature Genetics IF 41.307
Research results: Cervical cancer is the most common malignancy of the female reproductive system, and human papillomavirus (HPV) integration is a key event in cervical carcinogenesis. By performing genome-wide HPV integration profiling in cervical cancer (CC), the team identified clusters of genomic loci and revealed a potential mechanism of microhomology-mediated integration. In more than 100 samples, 3,667 HPV integration loci were identified, in addition to some common integration loci: POU5F1B (9.7%), FHIT (8.7%), KLF12 (7.8%), KLF5 (6.8%), LRP1B (5.8%) and LEPREL1 (4.9%). Some novel loci were also identified: HMGA2 ( 7.8%), DLG2 (4.9%) and SEMA3D (4.9%). These identified integration loci can be used as biomarkers for early personalized treatment and prognostic assessment of CC. Furthermore, FHIT and LRP1B protein expression was down-regulated when HPV was integrated into the FHIT and LRP1B introns, while MYC and HMGA2 protein expression was elevated when HPV was integrated into the flanking regions. In addition, the microhomology sequence between the human and HPV genomes was found to be significantly enriched near the integration site, suggesting that fusion between viral and human DNA may occur through a microhomology-mediated DNA repair pathway.

Molecular typing of gastric cancer can be used to identify different patient prognostic outcomes
Published: April 20, 2015
Publication: Nature Medicine IF 87.241
Research Results: Gastric cancer (GC) is one of the most deadly cancers and is highly heterogeneous. The research team combined GC heterogeneity characteristics and gene expression data to establish four molecular subtypes associated with different patterns of molecular alterations, disease progression and prognosis: MSI (microsatellite instability) subtype, MSI/EMT (microsatellite instability/epithelial-mesenchymal transition) subtype, MSS (microsatellite stability)/TP53+ subtype and MSS/TP53- subtype, which can help develop more customized treatment plans for different patients. The MSI/EMT subtypes can help develop more rational and personalized treatment plans for different patients. Among them, MSI/EMT has the worst prognosis, tends to occur in young patients and has the highest frequency of relapse (63%) among the four subtypes. The MSI subtype is common in the sinus and pylorus (70%) and often has an intestinal subtype (60%), with the best overall prognosis and lowest relapse frequency (22%). Both the MSS (microsatellite stable)/TP53+ subtype and the MSS/TP53- subtype have an intermediate prognosis and relapse rates. In addition, EBV virus infection rates were higher in the MSS/TP53+ subtype than in the other three groups.

Transforming FGFR-TACC gene fusion induces malignant glioblastoma
Published: September 7, 2012
Publication: Science IF 63.714
Research results: Glioblastoma multiforme (GBM), a brain tumor, is one of the most deadly cancers in humans. The team found that a small proportion of GBMs (3.1%; 3 of 97 tumors examined) contain oncogenic chromosomal translocations that allow the tyrosine kinase-encoding domains (FGFR1 or FGFR3) of the fibroblast growth factor receptor (FGFR) gene to fuse with TACC1 or TACC3, respectively, which encode the translesion acidic convoluted helix (TACC) structural domain. The FGFR-TACC fusion protein exhibits oncogenic activity when introduced into astrocytes or stereotactically transduced in a mouse brain. At the same time, it has constitutive kinase activity when localized at both poles of the mitotic spindle, inducing mitotic and chromosomal segregation defects, thereby triggering aneuploidy. Inhibition of FGFR kinase activity corrects aneuploidy, so oral FGFR inhibitors prolong the survival time of intracranial FGFR3-TACC3-induced glioma mice. The FGFR-TACC fusion is expected to be a potential predictor of whether some GBM patients will benefit from treatment with targeted FGFR kinase inhibitors.

Genomic basis and genetic mechanisms of transcriptomic changes in pan-cancerous species
Published: February 5th, 2020
Publication: Nature IF 69.5
Research results: Transcriptional alterations are usually caused by somatic changes in the cancer genome. During cancer development, multiple modes of RNA alteration are covered, such as overexpression, altered splicing and gene fusion. However, it is difficult to correlate these with underlying genomic changes due to the heterogeneity between patients and tumor types. The team used matched whole-genome sequencing data to correlate multiple RNA alterations with germline and somatic DNA alterations to propose a catalog of cancer-associated genetic alterations across 27 different types of cancer. The study identified 1,900 splicing alterations associated with somatic mutations; it also confirmed that 82% of gene fusions were associated with structural variants. Moreover, transcriptomic alterations came with different characterizations in each different cancers, but were all associated with changes in DNA mutational features. This study of RNA alterations in a genomic context provides a heavy basis for discovering genes and genetic mechanisms associated with cancer function.
About BGI Genomics
BGI Genomics, headquartered in Shenzhen China, is the world’s leading integrated solutions provider of precision medicine. Our services cover more than 100 countries and regions, involving more than 2,300 medical institutions. In July of 2017, as a subsidiary of BGI Group, BGI Genomics (300676.SZ) was officially listed on the Shenzhen Stock Exchange.BGI has topped the Asia Pacific and China life science corporate institution ranking table for the seventh year running, released in the 2022 Nature Index Annual Tables.