Service Description
BGI offers multi-omics services to look across genomics, transcriptomics, epigenomics, proteomics and metabolomics, with the flexibility to customize solutions that meet your specific needs. All projects are supported by an advanced bioinformatics infrastructure.
Metabolome + Genome re-sequencing Correlation Analysis(mGWAS)
Early correlation studies only focused on individual and highly heritable macrophenotypes, with the aim to identify a few common major gene variants. Fast advances in high-throughput sequencing technologies, have allowed scientists to expand their targets to rare variants on a genome-wide scale and focus on elucidating the genetic basis of complex human diseases, or quantitative traits in animals and plants.
Metabolomics, based on high-throughput mass spectrometry, can decompose a small number of macroscopic phenotypes into metabolic molecular phenotypes, and find significantly associated metabolic indicators and gene variants through mGWAS, which can more directly reveal the molecular mechanism behind macroscopic phenotypes such as diseases. As a research hotspot in recent years, mGWAS has been widely used in animal and plant variety breeding, human disease research and other fields1-4.
Project Workflow
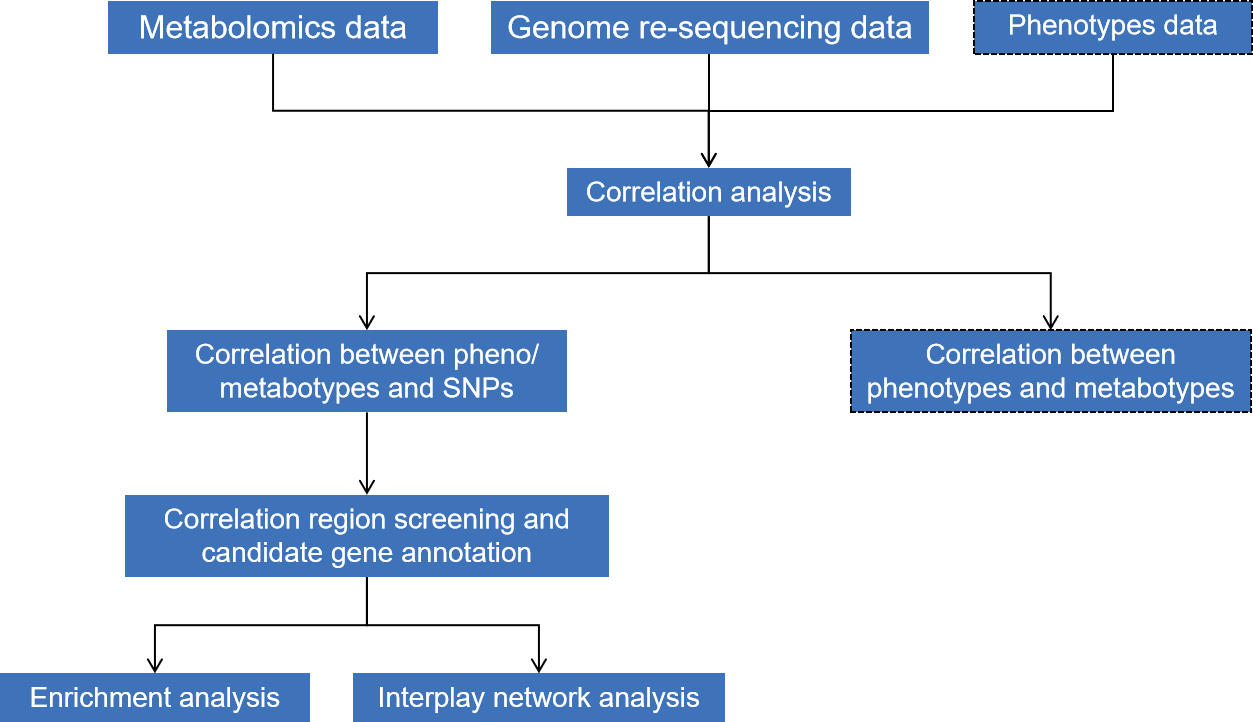
Note: The virtual box is optional
References
[1] Zhou Y, Zhang Z. et al. Graph pangenome captures missing heritability and empowers tomato breeding. Nature. 2022 Jun; 606(7914): 527-534.
[2] Cheng Y, Schlosser P. et al. Rare genetic variants affecting urine metabolite levels link population variation to inborn errors of metabolism. Nature Communications. 2021 Feb 11; 12(1): 964.
[3] Wang X, Kadarmideen HN. Metabolite genome-wide association study (mGWAS) and gene-metabolite interaction network analysis reveal potential biomarkers for feed efficiency in pigs. Metabolites. 2020 May 15; 10(5): 201.
[4] Zhu G, Wang S. et al. Rewiring of the fruit metabolome in tomato breeding. Cell. 2018 Jan 11; 172(1-2): 249-261.
How to order
Mass Spectrometry Service Specification

Quality Standard
- Summary includes data analysis
- Reports provided in PDF format, RAW files available upon request

Turn Around Time
- Typical 2-4 weeks from LC-MS/MS raw data acceptance to data report delivery
Sample Requirements
| INTEGRATIVE ANALYSIS | TOTAL NUMBER OF SAMPLES | BIOLOGICAL DUPLICATES |
|---|---|---|
| Genome re-sequencing+ metabolome | > 100 | ≥ 10 |
Data Analysis
- Correlation between pheno/ metabotypes and SNPs
- Correlation region screening and candidate gene annotation
- Correlation between phenotypes and metabotypes