Plant and Animal De Novo Genome Sequencing
De novo sequencing refers to the sequencing of a novel genome without a reference sequence for alignment. The process of de novo genome sequencing involves the sequencing of small/large DNA fragments, assembling the reads into longer sequences (contigs) and finally ordering the contigs to obtain the entire genome sequence.
BGI Genomics is a recognized leader in De Novo Genome Sequencing and has extensive experience from the de novo sequencing and assembly of more than 400 species’ genomes. We offer a complete suite of technologies to support your de novo sequencing projects, along with expert assistance in the planning of optimal sequencing and bioinformatics options, to ensure your project is a success.
Project Workflow
- SAMPLE PREPARATIONSample QC
- LIBRARY CONSTRUCTIONLibrary QC
- SEQUENCINGSequencing QC
- RAW DATA OUTPUTData QC
- BIOINFORMATICS ANALYSISDelivery QC
Sequencing Strategy
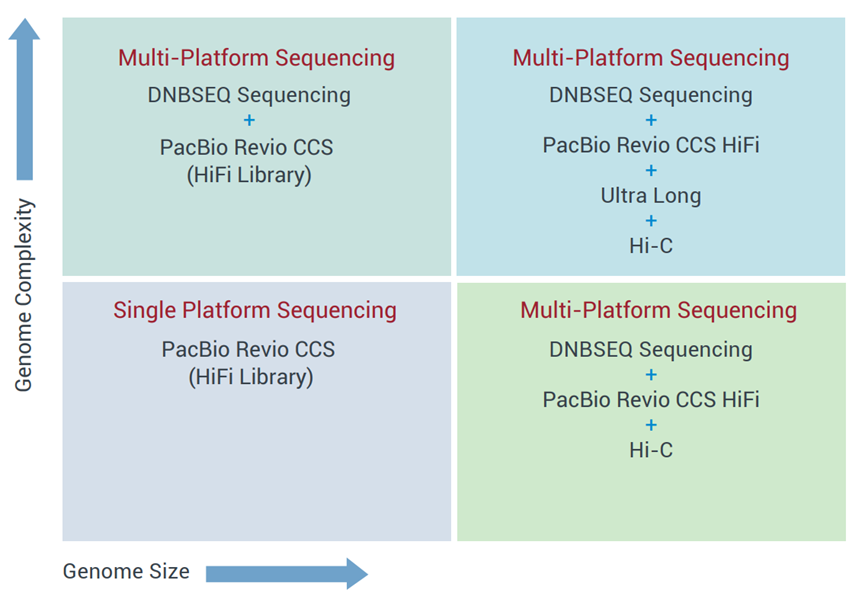
De novo sequencing usually requires a customized approach based on your subject species’ genome size and complexity as well as the overall scientific objectives of the project.
Our plant and animal de novo sequencing services are usually performed using a combination of available platforms, including proprietary DNBSEQ™ platforms augmented with Nanopore, PacBio Revio for sequencing, library preparation and mapping. In addition, we offer extensive bioinformatics data analysis options for genome assembly, annotation and evolution studies.
| Platform Tools | Library type | Sequencing | Recommend sequencing depth |
|---|---|---|---|
| DNBSEQ | 350bp Library | PE150 | ≥100X |
| Hi-C Library | PE150 | ≥100X | |
| Nanopore PromethION | 20-40Kb Library | Read length ≥10Kb | 30-100X |
| Ultra long library(>50K) | Read length ≥20Kb | 30-100X | |
| PacBio Revio | 15Kb CCS(HIFI) Library | Read length ≥10Kb | 30-60X |
Our Sequencing specialists will work with you to design the optimal strategies for your project, using platform combinations as appropriate for your project.
How to order
Sequencing Service Specification

Sample preparation and services
- Library preparations (DNBSEQ™, Nanopore, PacBio Revio etc)
- Various sequencing platforms
- Raw data, standard and customized data analysis
- Available data storage and bioinformatics applications

Turn Around Time
- Nanopore (normal library): Typical 30 working days from Sample QC acceptance to data analysis report availability
- PacBio: Typical 30 working days from Sample QC acceptance to data analysis report availability
- Case by case for the genome analysis
- Expedited services are available; contact your local BGI Genomics specialist for details

Sequencing quality standard
- Guaranteed ≥85% of DNBSEQ™ clean bases with quality score of Q30
- Guaranteed ≥80 Gb PacBio Revio HiFi data except for some complex species
Sample Requirements
| Platform | Library Type | Mass | Concentration | OD | Integrity (AGE) |
|---|---|---|---|---|---|
| DNBSEQ | 350 bp library | ≥0.2 μg (recommend≥0.4 μg) | 8 ng/μL | - | The band shown on gel electrophoresis has little degradation, or of fragment size greater than 20 kb. |
| Nanopore | 20-50 Kb Normal long library | ≥2 μg | 50 ng/μL | OD260/280: 1.8-2.0 OD260/230:≥1.5 | The band shown on gel electrophoresis has no or little degradation. |
| Nanopore | Ultra long library | ≥10 μg | 50 ng/μL | OD260/280: 1.8-2.0 OD260/230:≥1.5 | The band shown on gel electrophoresis has no or little degradation. |
| PacBio Revio | 15-20 Kb HIFI library | ≥14 μg | 80 ng/μL | OD260/280: 1.6-2.2 OD260/230: 1.6-2.5 | The band shown on gel electrophoresis has no or little degradation. (The main peak is around 40 Kb with smear no smaller than 20 Kb) |
Data Analysis
| Service Content | |
| Genome Survey | 1.Kmer estimation (Jellyfish + GenomeScope); |
| Genome Assembly (Pacbio HiFi data) | 1. Assembly; 2. Assessment by short reads alignment; 3. BUSCO assessment ; |
| Gene Annotation | 1.Repeat annotation; 2.Gene structure annotation; 3.Gene function Annotation; |
| Evolution | Deliver published genome and allied species (less than 10 species) 1.Gene family identification (Animal TreeFam; Plant OrthoMCL;≤10 species); 2.Phylogenetic tree construction; 3.Estimation of divergence time; 4.Genome synteny analysis; 5.Whole genome duplication analysis; 6.Gene family expansion and contraction analysis; |
| Auxiliary Assembly | Hi-C data auxiliary assembly |
CUSTOM ANALYSIS
Further customization of Bioinformatics analysis to suit your unique project is available: Please contact your BGI Genomics technical representative.

