Plant & Animal T2T Genome
Plant & Animal T2T Genome Introduction
The telomere-to-telomere (T2T) genome refers to a high-quality complete genome with high genomic accuracy, high continuity, and high integrity. This is conducive to the in-depth study of highly repetitive sequence regions in the genome and helps to resolve centromeres variation characteristics and evolutionary patterns of complex structures such as telomeric and telomeres.
The continuous optimization of combined assembly algorithms has greatly facilitated the De novo assembly of genomes. In more and more species, assembly gaps caused by complex regions of the genome, complex sex chromosomes, and haploid genomes in the polyploid assembly have been analyzed [1-4]. These findings have implications for disease occurrence and species evolution. Therefore, the construction of a "perfect reference genome" containing T2T, haplotype information, and sex chromosome information has become a research trend in genomics.
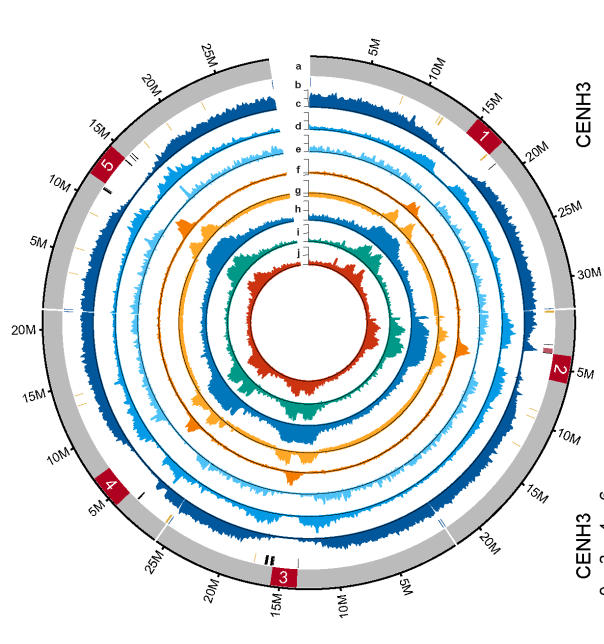
Figure 1. Complete map of the Arabidopsis genome. [4]
T2T genome research application features:

Resources
References
[1] Miga, K. H. et al. Telomere-to-telomere assembly of a complete human X chromosome. Nature 585, 79–84 (2020).
[2] Logsdon, G. A. et al. The structure, function and evolution of a complete human chromosome 8. Nature 593, 101–107 (2021).
[3] Nurk, S. et al. The complete sequence of a human genome. Science 376, 44–53 (2022).
[4] Naish, M. et al. The genetic and epigenetic landscape of the Arabidopsis centromeres. Science 374, eabi7489 (2021).